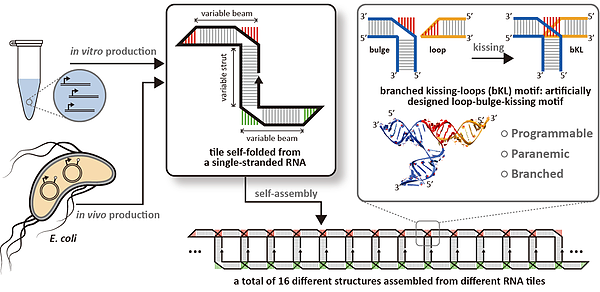
In a study published last week in Nature Chemistry, Prof. Yossi Weizmann of BGU's Department of Chemistry and colleagues unveiled an innovative approach to push the limits of making custom homooligomerization made from RNA tiles, which allows them to interact via an artificial branched kissing-loop (bKL). The new motif facilitates the design of numerous homooligomerizable tiles that are folded from a single-strand of RNA and assemble into diverse structures with specific shape and size comparable to those of natural biological systems.
Homooligomerization is the self-assembly of a single species into a macrostructure and is a ubiquitous strategy in biological systems. It enables cells to create different and complex geometries with exceptional efficiencies, such as in the case of actin and tubulin cytoskeletons. However, to design artificial nanostructures comparable to those of natural systems is very challenging. The answer was to program the RNA single-stranded into various monomer tiles, which, because cells are naturally suited to generating large amounts of single-stranded RNAs, in vivo expression and large-scale production of artificial structures can be made readily.
This work will be of broad interest to researchers beyond the communities of structural RNA or DNA nanotechnology. The new artificial motif displays similarity to motifs in natural functional RNAs; therefore, it can help to those investigating natural RNA structures. Furthermore, the expression of RNA tiles based on the bKL motif in living cells, and confirmed its tolerance to functional aptamers, can help to the wider synthetic biology community, especially for the researchers that engineering RNAs within cells.