In the last decade, proteomics has emerged as a promising field for studying global gene expression profiles at the protein level. Developments in mass spectrometry-based technologies enable the identification of proteins and measurement of their abundance in biological samples. This approach, often termed "quantitative proteomics", provides a useful platform for the study of quantitative differences in protein abundance among conditions, tissues or cell types.
Initial preprocessing and analysis of the mas-spec data is usually carried out at the proteomics laboratory, yielding an excel file specifying estimated abundance per protein per sample.
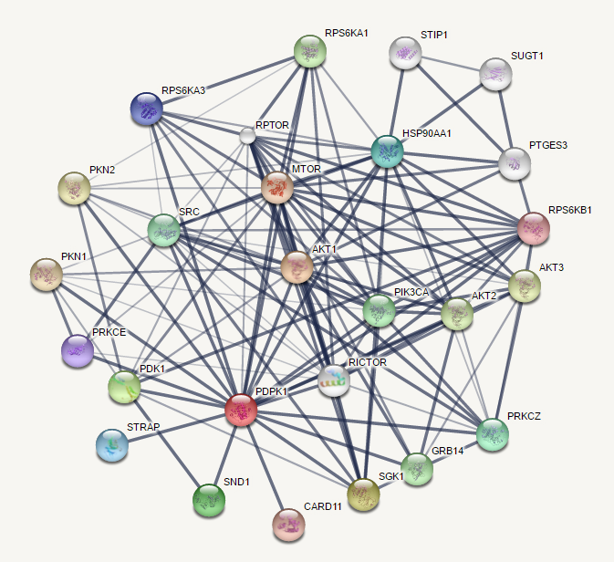
Subsequent statistical tests for comparing protein abundance among the biological treatments or states, can then be performed at the Bioinformatics Core Facility, along with downstream analyses such as clustering, functional and pathway enrichment.
So far, we have analyzed numerous mass-spec proteomics profiling datasets, using Partek(R) Genomics Suite, Perseus and in-house R programs.