Bacterial colonies spread rapidly on surfaces by collective motion. This motion, termed “bacterial swarming,” is intricate and is similar in its dynamics to schools of fish and flocks of birds. Thus, this phenomenon has vast physical, biological and ecological importance.
Most of the works on collective bacterial motion have focused only on thin colonies in which a very few layers of bacteria move one on top of the other. This was done mainly due to technical difficulties in tracking cells growing in a thick, three-dimensional, and perhaps more realistic structure. In thin colonies, introductions of antibiotics to the colonies eliminate the cell population almost instantaneously.
Recent research carried out by Prof. Avraham Be’er from the Zuckerberg Institute for Water Research at Ben Gurion University, together with Prof Gil Ariel from the Department of mathematics at Bar Ilan University, and Prof Rasika Harshey from Microbiology at The University of Texas at Austin studied the dynamics and geometry of three-dimensional swarming colonies. By using phase contrast imaging, combined with off-focus fluorescent imaging, the time-resolved 3D geometry of the colony was resolved.
The analysis reveals a novel 3D architecture, where bacteria are constrained by inter-particle interactions by being sandwiched between two distinct boundary conditions. It was found, that secreted biosurfactants keep bacteria away from the swarm-air upper boundary, and added antibiotics at the lower swarm-surface boundary lead to their migration away from this boundary. “The formation of the antibiotic-avoidance zone,” says Prof. Be’er, “is dependent on a functional chemotaxis signaling system, in the absence of which the swarm loses its high tolerance to the antibiotics. Therefore, it is clear that the cells migrate to safer regions of the colony and that the 3D structure of the colony assists the strain in surviving adverse conditions”.
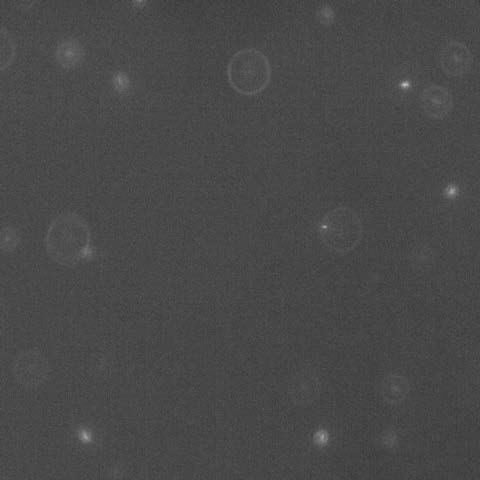
In the Photo: a snapshot of an off-focus fluorescent image viewed from the top. The diameters of the diffraction rings report on bacterial location at different heights in the colony. Cells in focus are on the agar. As an example, the larger ring in the picture corresponds to the location of a cell 35 μm above the agar. There are, on average, 1000 times more unlabeled, nonfluorescent, cells in any view of the fluorescence image as determined by viewing the same field in phase contrast microscopy